Application of Bioinformatics & Systems Analyses Approaches
We have established meta-analyses system biology pipelines for meaningful interpretation of complex biological process. Utilizing these pipelines we have integrated multi-layer datasets and have identified novel genes, regulators, pathways and small molecules associated with various biological questions.
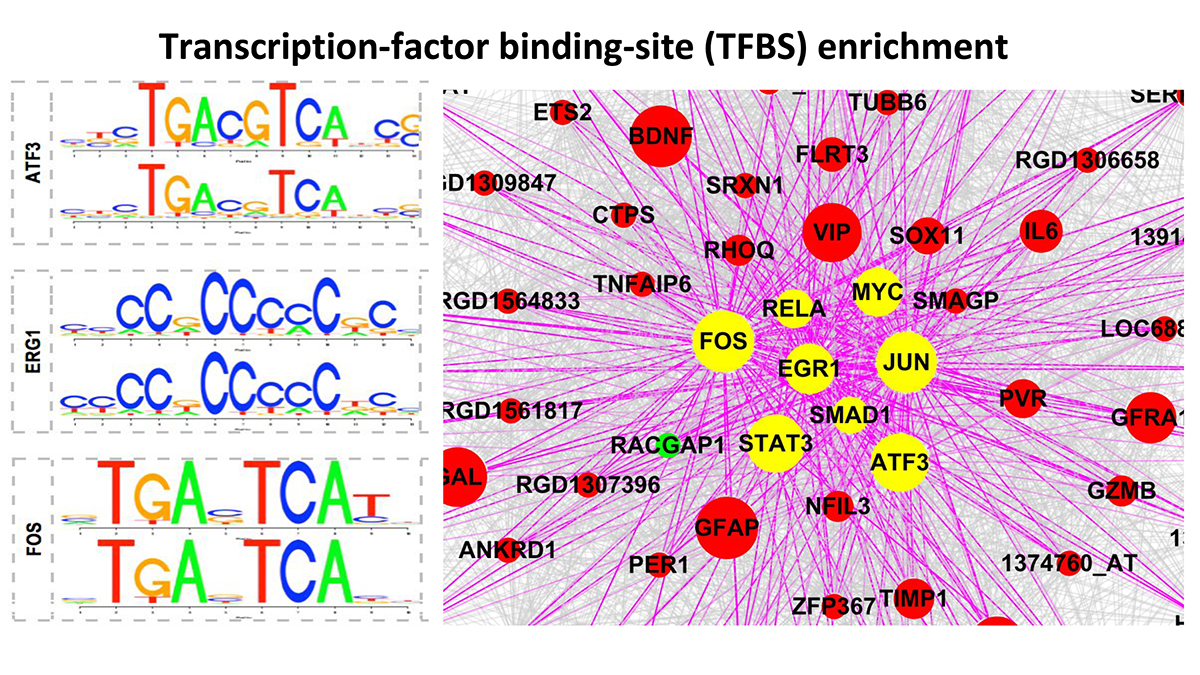
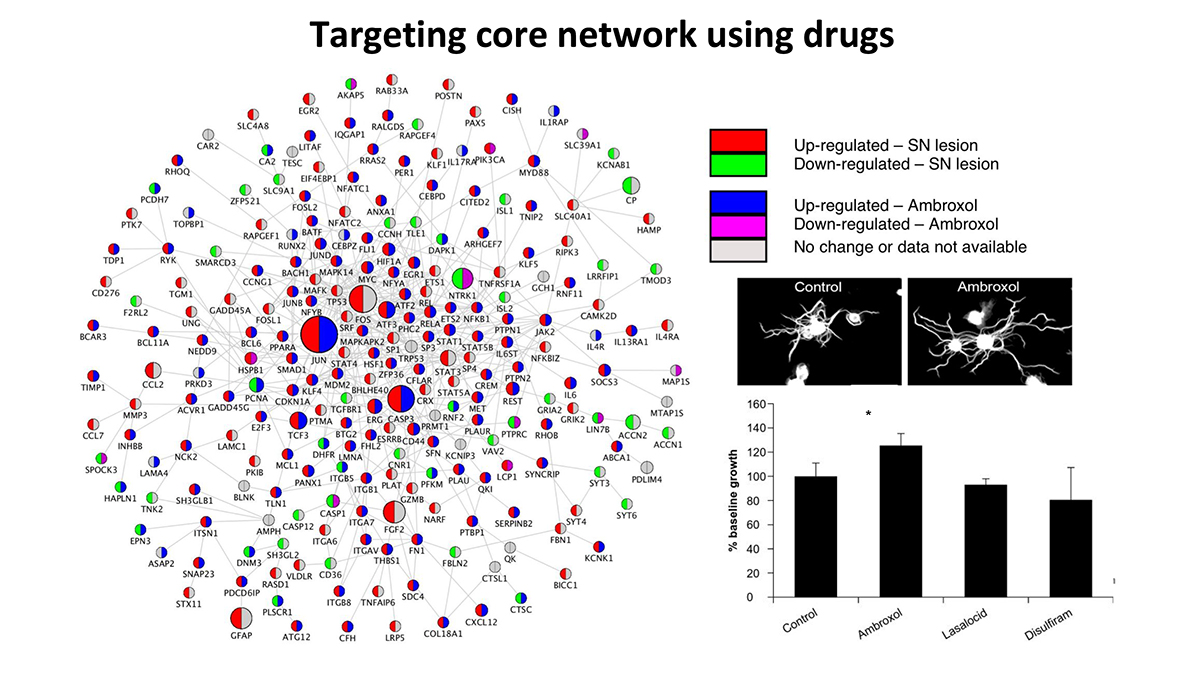
By applying these pipelines to nerve injury datasets, we have identified, (1) a transcriptional program observed after PNS, but not CNS injury, (2) this program links known signaling pathways via a core set of transcription factors, (3) we experimentally and bio-informatically validated several network predictions and (4) used the core transcriptional profile to identify a drug that promotes CNS regeneration.
By applying these approaches to transcriptomic studies obtained from autism spectrum disorder (ASD) samples, we have demonstrated the presences of pathway convergence and identified critical transcriptional regulators associated with ASD. We have also applied genomic, proteomics and metabolomics framework to understand evolutionary process in Escherichia coli, and demonstrated how a network framework facilitates hypothesis generation and permits evolving beyond network predictions to experimental validation of network models.